Chemicals and reagents
NG,NG´-dimethyl-L-arginine di(p-hydroxyazobenzene-p´-sulfonate) (SDMA), S-adenosyl-L-methionine (SAM), n-butyryl-L-carnitine (nC4), iso-butyryl-L-carnitine (iC4) and citrulline (CIT) were obtained from Sigma-Aldrich (Steinheim, Germany). Creatinine (CNN) was provided by Alfa Aesar (Karlsruhe, Germany). D-erytro-sphingosine-1-phosphate (S1P) was bought from Larodan AB (Limhamn, Sweden). Cis-4-decenoylcarnitine was synthesized by Lumila research group at the Autonomous University of Madrid (Madrid, Spain). Bilirubin (BIL) was purchased from TCI (Tokyo, Japan). Isotopically labeled compounds, NG,NG´-dimethyl-L-arginine-d6 (SDMA-d6) and creatinine-d3 (CNN-d3) were supplied by Toronto Research Chemicals, TRC-Canada (North York, Canada). Acetonitrile was obtained from Scharlau (Sentmenat, Spain), LC–MS grade formic acid from FisherScientific (Ghent, Belgium), ammonium formate from Sigma-Aldrich (Steinheim, Germany) and perfluoroheptanoic acid 96% (PFHA) from Acros Organics (New Jersey, USA). The mobile phase was filtered through 0.1 μm filters from Millipore Omnipore (Watford, Ireland). Acetic acid was supplied by Fisher Scientific (Loughborough, UK).
Study samples
Blood-derived samples were collected from sixty-seven CKD children and forty-five healthy pediatric volunteers from two countries, as described below.
We employed the Schwartz formula to define CKD in our study. The Schwartz formula utilizes serum creatinine levels, height, and an empirical constant to estimate the Glomerular Filtration Rate (GFR). This formula has been widely accepted and utilized as an enrollment criterion in various studies, including the Chronic Kidney Disease in Children study. This National Institutes of Health–funded North American cohort study aims to recruit children and adolescents with mild to moderate CKD. Its primary objectives include characterizing disease progression and assessing the impacts of CKD on cardiovascular health, growth parameters, and behavioral indices.
Plasma samples were collected at Cruces University Hospital in Barakaldo (Basque Country, Spain). Blood samples from thirty-one CKD patients, aged 3–17 (average age: 10.9 years old), and thirteen healthy control patients aged 6–16 (average age: 9.8 years old). The study protocol was approved by the Ethics Committee of Clinic Research of Cruces Hospital (approval number: E08/62). Patients and patients’ caregivers all provided informed consents. The study was performed in compliance with Spanish law on biomedical research (Law 14/2007, of July 3, on Biomedical Research, BOE no159, pp28826–28,848). Blood was drawn in the morning after overnight fasting in Beckton Dickinson EDTA tubes (Plymouth, UK). After centrifugation at 1000 g for 5 min at 4 °C, plasma samples were stored at − 80 °C until further analysis.
Serum samples were collected at the Department of Pediatrics and Nephrology of the Medical University of Bialystok (Podlasie Province, Poland). Blood samples were obtained from thirty-six CKD pediatric patients aged from 3 months to 17 years old (average age: 11.6 years) and thirty-two healthy controls aged from 1 week to 17 years old (average age: 5.9 years). The study was approved by the Institutional Review Board of the Medical University of Bialystok (R-I-002/301/2019). Blood was drawn in the morning after overnight fasting in Beckton Dickinson serum tubes (Plymouth, UK). Blood samples were left to clot and then centrifuged at 1000 g for 5 min at 4 °C. Serum samples were stored at − 80 °C until further analysis.
The exclusion criterium in both control and study groups was the occurrence of systemic (e.g., metabolic, autoimmune, neoplastic, and infectious) diseases. Children taking non-steroidal anti-inflammatory drugs (NSAIDs), glucocorticosteroids, hormones, antibiotics, vitamins, and dietary supplements for at least three months before the study were also excluded.
Sample processing
After thawing at room temperature, 50 µL of a sample was placed in Eppendorf tubes and spiked with 10 µL of a mix solution containing 100 µg mL−1 of creatinine-d3 and 1 µg mL−1 of SDMA-d6. Then, protein precipitation was performed by adding 150 µL of cold acetonitrile, vortexing the mixture, and centrifuging for 10 min at 15,600 g at 4 °C. The obtained supernatant was transferred to chromatographic vials and evaporated in a nitrogen stream in a Techne, Dri-Block®DB-3D (Staffordshire, UK) evaporator system. Finally, the residue was reconstituted in 100 µL of acetonitrile: acetic acid 0.5 M (75:25; v:v) solution and 2 µl were analyzed using LC-QQQ-MS.
LC-QQQ-MS method
Chromatographic analysis of blood samples was performed on an Agilent 1200 Series HPLC system coupled to Agilent 6410 Series triple quadrupole mass spectrometer (LC-QQQ-MS) from Agilent Technologies (Santa Clara, CA, USA), equipped with an electrospray source (ESI). Chromatographic separation was carried out in a Zorbax Eclipse Plus C18 (3.0 × 50 mm, 1.8 µm) reversed-phase column preceded by a C8 guard column (2.1 × 12 mm, 5 µm), both from Agilent Technologies. The mobile phase composition, gradient program, and column temperature were published previously20. Mass spectrometry measurements were recorded in the following conditions: fragmentor voltage, 100 V; drying gas temperature and flow, 250 °C and 10 L/min; nebulizer pressure, 40 psig; and capillary voltage, 3500 V. Quantification was performed by dynamic multiple reaction monitoring (dMRM) mode. Retention time, 1 precursor ion, and 2 product ions were used to identify and quantify the analytes in the samples20. iC4 and its structural isomer nC4 present the same ionization pattern in MS and MS/MS modes, thus it was necessary to include both isomers in the method to ensure the correct identification and quantification of nC4. This analytical method was previously evaluated in terms of linearity, trueness, precision, limits of quantification, and stability, as described elsewhere20.
Agilent Mass Hunter Workstation Data Acquisition version B.08.00 software and Mass Hunter Qualitative Analysis version B.07.00, both Agilent Technologies, were used for data acquisition and raw data processing. Data analysis was carried out using SPSS Statistics 23 from IBM (Armonk, New York, USA), Metaboanalyst, and Matlab R2015a (Mathworks, Natick, Massachusetts, United States).
Data analysis
Univariate data analysis was carried out to find whether differences in biomarkers concentration between CKD and control groups were significant. The distribution of blood concentration of each analyte in control and CKD patients was evaluated with the Kolmogorov–Smirnov test. The student’s t-test was applied for parametric variables, whereas the Mann–Whitney U test was applied to non-parametric variables.
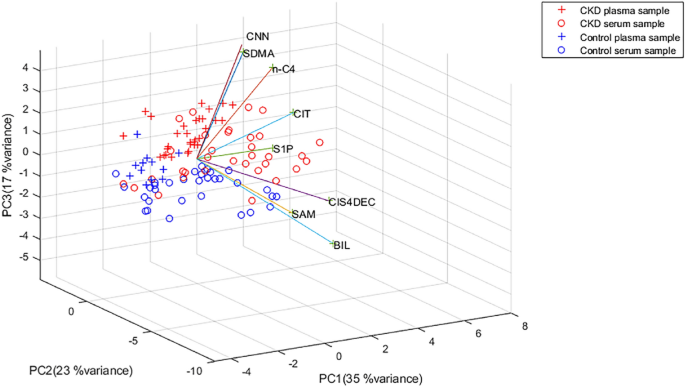
The intrinsic interdependency of the metabolite concentrations was evaluated by multivariate data analysis. Logarithm transformation of data was performed to analyze the data set in a normal distribution range. Subsequently, due to different concentration ranges of metabolites, data was Pareto scaled by normalizing the concentration of each analyte, subtracting the mean metabolite concentration, and dividing it by the square root of its standard deviation. Then, a PCA model was built for all samples.
The predictive ability of the proposed biomarkers was analyzed by ROC curves that describe the relationships between the sensitivity and specificity. Classical univariate ROC curve analysis was performed for CNN alone. In addition, multivariate ROC curves were generated using the PLS-DA algorithms for building the classification method using two-thirds of the samples and validating the approach on the 1/3 samples left out.
All data (concentration of the analytes) were log transformed before data normalization by Pareto scaling.
Institutional review board
The protocol of the study was approved by the local Ethical Committee of Clinic Research of Cruces Hospital (approval number: E08/62) and the Medical University of Bialystok, Poland (number R-I-002/301/2019).
- The Renal Warrior Project. Join Now
- Source: https://www.nature.com/articles/s41598-024-62518-w
